
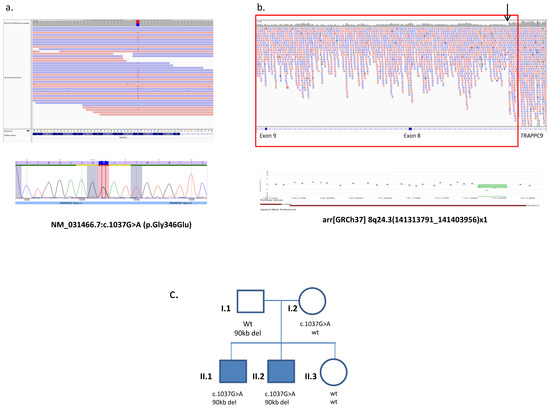
Amplification of EGR2 amplicons 2a, 2b, and 2d required the addition of Qiagen Q buffer to 20%. Using 50 ng of patient genomic DNA, the appropriate primers and Qiagen HotStarTaq, each region was amplified as follows: 15 minutes at 95☌, 40 cycles of amplification (95☌ for 30 seconds, 65☌ for 30 seconds, 72☌ for 1 minute), and 7 minutes at 72☌. All forward primers had a −21 M13 primer tail (TGTAAAACGACGGCCAGT), and all reverse primers had an M13 reverse tail (CAGGAAACAGCTATGACC). Primers for PCR amplification of exons and intronic splice junctions were designed using the Primer v3 program ( ) ( Table 1). Under the optimized DHPLC conditions, we report for screening these four genes, we detected two GJB1 mutations missed by automated DNA sequence analysis. To this end, we assembled a cohort of 168 CMT patient DNA samples that had tested negative for mutations by direct DNA sequencing and screened the DNA samples for PMP22, MPZ, GJB1, and EGR2 mutations by DHPLC. If this hypothesis were true, we reasoned that we could identify new mutations in the DNA samples from patients previously screened by direct sequencing. The supporting data of a heteroduplex peak in a DHPLC elution profile can be very helpful for focusing further sequencing efforts and scrutiny of the sequence chromatograms. Although sequencing has been the gold standard for mutation detection, data from sequence chromatograms are not always ideal and sequence analysis programs occasionally fail to detect heterozygous mutations.
In addition to improving the efficiency of mutation screening for CMT, we hypothesized that detection of a heteroduplex by DHPLC might improve the mutation detection rate for sequencing.


 0 kommentar(er)
0 kommentar(er)
